Submit parallel jobs on HPC
The workflow discussed in this post is particularly suited for scenarios in which you need to execute similar code multiple times, with each execution having a substantial time requirement. In each iteration, the code exhibits minor variations in specific input parameters. Then this is when the parallel computing comes into play.
Toy Task: Suppose I’d like to numerically evaluate the central limit theorem.
Central Limit Theorem: suppose \(\{X_1,X_2,\dots,X_n\}\) is a sequence of i.i.d. random variables with \(E(X_i)=\mu\) and \(Var(X_i)=\sigma^2 <\infty\) then as \(n\rightarrow\infty\), \(\bar{X}\rightarrow N(\mu,\frac{\sigma^2}{n})\)
Write a R function that perform the task
Create a R script, named task.R that simulate \(X\) with my specified sample size and seed, and output the sample mean of the simulated \(X\) values.
The following code takes two command-line arguments n and seed and output the sample mean to a Rdata file.
args = commandArgs(trailingOnly=T)
n=args[1]
seed=args[2]
set.seed(seed)
# generate data
x <- rnorm(n,0,1)
# sample mean of X
E.x <- mean(x)
# save the output
save(list = c("E.x"),file = paste0("output_",n,"_",seed,".Rdata"))
Note:* How to create the task.R file in linux?
- Create a file named task.R with the following code
vim task.R
- type
ito enter the insert mode, and paste the above code. - press
Escto exit the insert mode, and type:wqto save and quit
Upon creating the task.R file, we currently have one file in folder

To test the code, let \(n=100\) and \(seed=10\). Run the following code:
# load R
module load R
# run the job
Rscript task.R 100 10
It will produce a file named output_100_10.Rdata . Now we have two files in folder
Generate the jobs
We want to execute the task.R script for different sample sizes: 250, 500, 1000, 2000, and 4000. For each sample size, we intend to run task.R 1000 times by setting seeds from 1 to 1000.
For computational efficiency, we aim to run the \(5\times 1000\) jobs in parallel. To achieve this, first create a .txt file that contains all the jobs with the following R code. For reproducibility, I’d like to write the following code in a file called write_job.R. We now have three files in folder

# sample size
n.vec <- c(250,500,1000,2000, 4000)
# number of simulations
nsim <- 1000
for (i in seq_along(n.vec)){
joblist <- c()
for (t in 1:nsim){
job <- paste0("Rscript task.R ",n.vec[i]," ",t)
joblist <- c(joblist,job)
}
write.table(joblist, file = paste0("joblist_n",i,".txt"),quote = F, col.names = F, row.names = F)
}
Usually, we don’t want to submit too many jobs at a time, so I split the jobs into 5 files, named joblist_n250.txt, joblist_n500.txt, joblist_n1000.txt, joblist_n2000.txt, joblist_n4000.txt.
To generate those .txt files, type Rscript write_job.R in the terminal, which will execute the R code contained in the write_job.R file.

Submit parallel jobs
Personally I prefer to submit parallel jobs with the help of dSQ command.
- download dSQ (you will need to install git command first if it’s not already installed).
# Download dSQ
git clone https://github.com/ycrc/dSQ
# Define the path to the dSQ folder
dsq_path="/path/to/my/dSQdirectory"
- Generate bash jobs
module load python/3.8
module load R/4.2.2
##########################
# This is step1_dsq.sh
##########################
for i in $(seq 1 5); do
python "$dsq_path/dSQ.py" --job-file joblist_n$i.txt --batch-file joblist_n$i.sh --job-name n$i --mail-type ALL
done
- Modify the bash jobs to contain email and partition information. Change the partition name as need. I’m using the
day-long-cpupartition here.
##########################
# This is step2_modify.sh
##########################
for i in $(seq 1 5); do
# add "python" at the last line
sed -i 's@'"$dsq_path"'/dSQBatch.py@python '"$dsq_path"'/dSQBatch.py@' "joblist_n$i.sh"
# add email and packages
sed -i '/#SBATCH --mail-type "ALL"/a #SBATCH --mail-user=your-email@xxx.edu\nmodule load python/3.8\nmodule load R/4.2.2' joblist_n$i.sh
# specify partition name
sed -i '1a #SBATCH --partition=day-long-cpu' joblist_n$i.sh
done
Upon this step, we should have the following files in folder

- submit the jobs one by one, so we don’t abuse the cluster:)
sbatch joblist_n1.sh
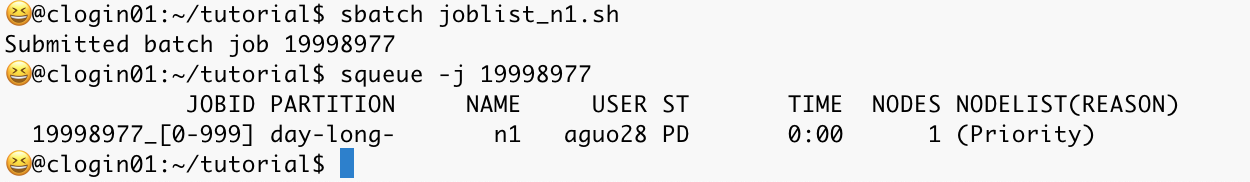
The above screenshot shows that we have successfully submitted the bash job joblist_n1.sh and the submission id is 19998977. squeue -j 19998977 helps check the current status of this job, and the PD in the ST means the jobs are queued and awaiting execution; they have not commenced running yet.
if we want to submit all the jobs at once:
##########################
# This is step3_submit.sh
##########################
for i in $(seq 1 5); do
sbatch joblist_n$i.sh
done
Check job status using squeue
Upon submitting parallel jobs, we can check the job status with the squeue command.
squeue # return status of all jobs of all users
Running the squeue command alone will display the status of all pending and running jobs across the entire cluster for all users. This provides an overview of the cluster’s current workload. By examining the output, you can get an idea of how many users have submitted jobs and how many of those jobs are actively running or waiting in the queue.
squeue --job <your_job_number> # return status of jobs under <your_job_number>
squeue -u <your_user_name> # return status of jobs under <your_user_name>
To narrow down the results returned by the squeue command and focus on specific jobs or jobs belonging to your user account, you can use the --job and -u options respectively.
All the above commands will return the status of jobs via job state codes. The table below summarizes the different status codes displayed by the squeue command and their corresponding meanings. (table source)
| Status | Code | Explaination |
|---|---|---|
| COMPLETED | CD | The job has completed successfully. |
| COMPLETING | CG | The job is finishing but some processes are still active. |
| FAILED | F | The job terminated with a non-zero exit code and failed to execute. |
| PENDING | PD | The job is waiting for resource allocation. It will eventually run. |
| PREEMPTED | PR | The job was terminated because of preemption by another job. |
| RUNNING | R | The job currently is allocated to a node and is running. |
| SUSPENDED | S | A running job has been stopped with its cores released to other jobs. |
| STOPPED | ST | A running job has been stopped with its cores retained. |
To obtain more detailed job status information and monitor the progress of jobs more effectively. Here are some additional options:
-
--long: This option provides more detailed job status information beyond the basic fields. -
--start: When used with this option,squeuewill return the estimated start time for pending jobs. This estimation is based on the current workload and resource availability in the cluster. Note that this estimation can be inaccurate, as new jobs with higher priorities or changes in resource availability can affect the actual start time of a pending job. -
--iterate=: This option allows you to automatically refresh the job status information every specified number of seconds. This feature is useful for monitoring job progress without manually re-running the command. To stop the automatic iteration, simply pressCtrl+C.
squeue --job <your_job_number> --long # return more detailed status info
squeue -u <your_user_name> --start # return an estimated start time
squeue -u <your_user_name> --start --iterate=<n_seconds> # return an estimated start time every n_seconds
squeue command has way more options than I can introduce here. The command below allows us to check all the options that squeue has to offer. It will open a help page that contains all the squeue options along with detailed explantation. Press q to exit this help page.
man squeue # check all options of squeue command
Cancel jobs using scancel
Sometimes we may want to cancel the submitted jobs. scancel is designed for this purpose and its usage is very similar to squeue
scancel <your_job_number> # cancel certain job
scancel <your_job_number1>, <your_job_number2>, <your_job_number3> # cancel multiple jobs
scancel -u <your_user_name> # cancel all jobs from certain user
Control queued and running jobs using scontrol
To suspend a currently running job, we can use the suspend command.
scontrol suspend <your_job_number>
This will pause the currently running job and we can resume the job with resume
scontrol resume <your_job_number>
To hold a job that is queued but not yet running, we can use the hold command
scontrol hold <your_job_number>
Different from suspend, this hold command can only be applied to jobs that is currently waiting in line and not run yet. Using the hold command will assign lowest priority to the job such that other jobs can be run first. To cancel this holding status, we can use the release command.
scontrol release <your_job_number>
Organize the results
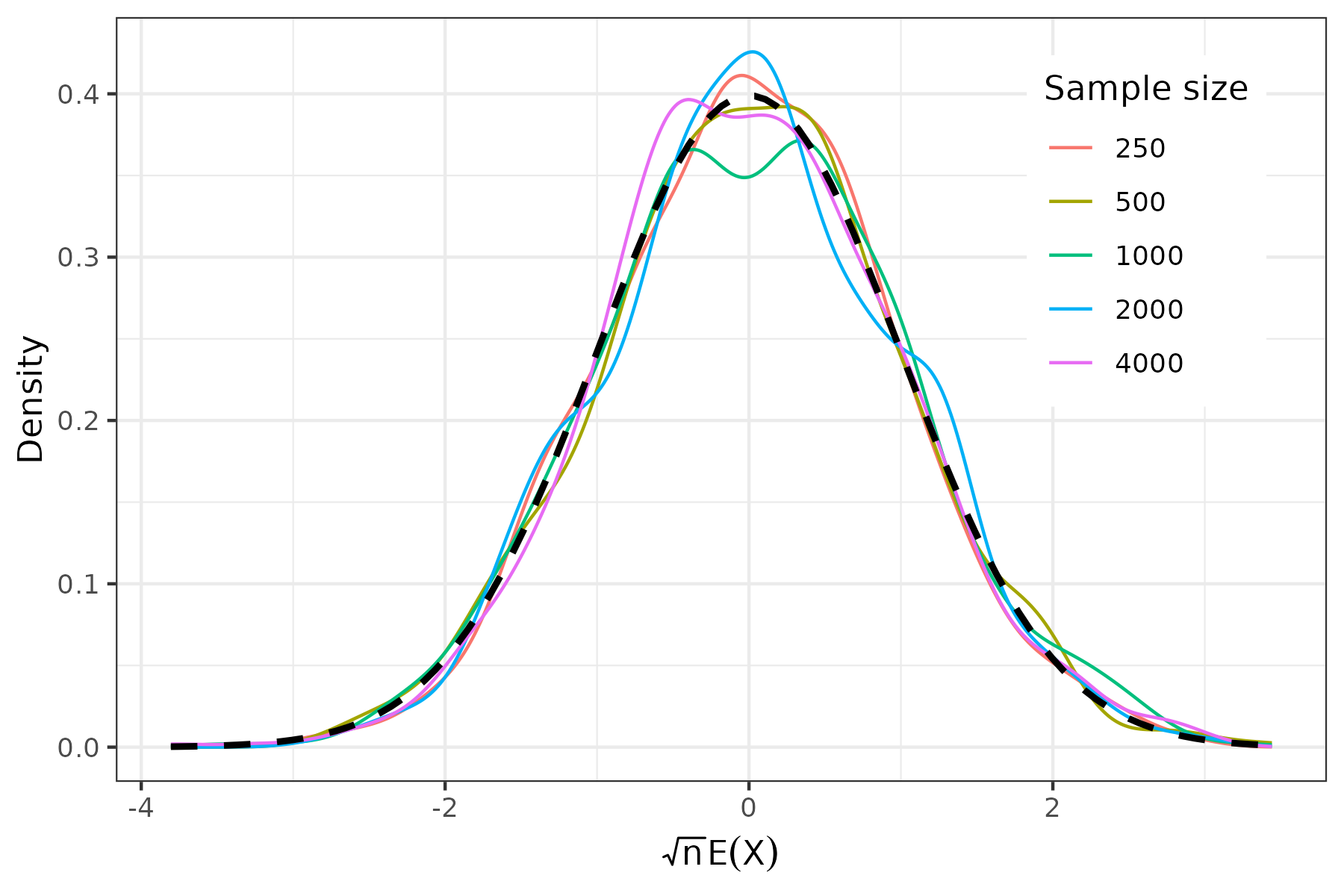
Finally, we are ready to organize the results. This step varies based on your task. In this toy example, we’d like to verify: \(\sqrt{n}(\bar{X}-0)\rightarrow N(0,1)\) Therefore, I’ll plot the kernel density estimate of \(\bar{X}\) for each sample size, and compare them with the standard normal distribution \(N(0,1)\)
The following R code does the job.
library(ggplot2)
library(dplyr)
library(latex2exp)
# sample size
n.vec <- c(250,500,1000,2000,4000)
# number of simulations
nsim <- 1000
# record results
mean_matrix <- matrix(nrow = nsim, ncol = length(n.vec))
for (i in seq_along(n.vec)){
# sample size
n <- n.vec[i]
for (t in 1:nsim){
load(paste0("output_",n,"_",t,".Rdata"))
# record bias
mean_matrix[t,i] <- E.x
}
}
# plot the result with boxplot
dt <- data.frame(n=rep(n.vec, each=nsim), value=c(mean_matrix)) %>% mutate(n.value=sqrt(n)*value)
######################
# density plot
######################
p.den <- ggplot(dt, aes(x=n.value, color=factor(n))) +
geom_density(key_glyph = draw_key_path)+
theme_bw()+
xlab(TeX(r'($\sqrt{n}E(X)$)')) +
ylab("Density")+
theme(legend.position = c(.95, .95),
legend.justification = c("right", "top"))+
guides(color = guide_legend(title = "Sample size"))
# Overlay a single normal curve
p.den <- p.den +
stat_function(
fun = dnorm,
args = list(mean = 0, sd = 1),
linetype = "dashed",
color = "black",
size = 1
)
# Save the ggplot visualization to a PNG file
ggsave(filename = "dens_plot.png", plot = p.den, device = "png", width = 6, height = 4, dpi = 300)
The resulting plot: